-Search query
-Search result
Showing 1 - 50 of 83 items for (author: lin & wc)

EMDB-41888: 
Structure of Apo CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41889: 
Structure of CXCL12-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41890: 
Structure of AMD3100-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41891: 
Structure of REGN7663 Fab-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41892: 
Structure of REGN7663-Fab bound CXCR4
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41893: 
Structure of trimeric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41894: 
Structure of tetrameric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC
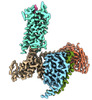
EMDB-36144: 
Senktide bound to active human neurokinin 3 receptor in complex with Gq
Method: single particle / : Sun WJ, Yang F, Zhang HH, Yuan QN, Yin WC, Shi P, Eric X, Tian CL

EMDB-36145: 
Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq
Method: single particle / : Sun WJ, Yang F, Zhang HH, Yuan QN, Yin WC, Shi P, Eric X, Tian CL
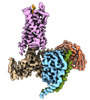
EMDB-36146: 
Substance P bound to active human neurokinin 3 receptor in complex with Gq
Method: single particle / : Sun WJ, Yang F, Zhang HH, Yuan QN, Yin WC, Shi P, Eric X, Tian CL

EMDB-42141: 
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WC, Eichman BF

EMDB-42142: 
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WC, Eichman BF

EMDB-37096: 
Rpd3S histone deacetylase complex
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, Jun H

EMDB-37122: 
Rpd3S in complex with 187bp nucleosome
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37123: 
Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37124: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37125: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2
Method: single particle / : Dong S, Li H, Meilin W, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37126: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37127: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Chang X, Chao WCH, He J

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41180: 
Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
Method: single particle / : Goldsmith JG, McLellan JS

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-33497: 
Neurokinin A bound to active human neurokinin 2 receptor in complex with G324
Method: single particle / : Sun WJ, Yuan QN, Zhang HH, Yang F, Ling SL, Lv P, Eric X, Tian CL, Yin WC, Shi P
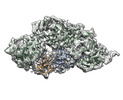
EMDB-40241: 
1:1:1 agrin/LRP4/MuSK complex
Method: single particle / : Xie T, Xu GJ, Liu Y, Quade B, Lin WC, Bai XC

EMDB-28570: 
CryoEM structure of PN45545 TCR-CD3 complex
Method: single particle / : Saotome K, Franklin MC

EMDB-28571: 
CryoEM structure of PN45545 TCR-CD3 in complex with HLA-A2 MAGEA4 (230-239)
Method: single particle / : Saotome K, Franklin MC

EMDB-28572: 
CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4
Method: single particle / : Saotome K, Franklin MC

EMDB-28573: 
CryoEM structure of HLA-A2 bound to MAGEA4 (230-239) peptide
Method: single particle / : Saotome K, Franklin MC

EMDB-28574: 
CryoEM structure of HLA-A2 bound to MAGEA8 (232-241) peptide
Method: single particle / : Saotome K, Franklin MC

EMDB-27221: 
Cryo-EM map of human LIF signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27227: 
Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from selected full particle dataset, with better resolution in the interaction core region
Method: single particle / : Zhou Y, Franklin MC

EMDB-27228: 
Cryo-EM structure of human CNTFR alpha in complex with the Fab fragments of two antibodies
Method: single particle / : Zhou Y, Franklin MC

EMDB-27229: 
Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from a particle subset, with lower global resolution but improved LIFR D6-D8 density
Method: single particle / : Zhou Y, Franklin MC

EMDB-27230: 
Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha
Method: single particle / : Zhou Y, Franklin MC

EMDB-27231: 
Cryo-EM map of human CLCF1 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27244: 
Cryo-EM map of detergent-solubilized human IL-6 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27246: 
Cryo-EM map of human IL-27 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27247: 
Cryo-EM map of human IL-27 signaling complex: focused refinement on the interaction core region
Method: single particle / : Zhou Y, Franklin MC

EMDB-34540: 
Type VI secretion system effector RhsP in its post-autoproteolysis and monomeric form
Method: single particle / : Tang L, Dong SQ, Rasheed N, Wu HW, Zhou NK, Li HD, Wang ML, Zheng J, He J, Chao WCH

EMDB-34541: 
Type VI secretion system effector RhsP in its pre-autoproteolysis and monomeric form
Method: single particle / : Tang L, Dong SQ, Rasheed N, Wu HW, Zhou NK, Li HD, Wang ML, Zheng J, He J, Chao WCH

EMDB-34542: 
Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form
Method: single particle / : Tang L, Dong SQ, Rasheed N, Wu HW, Zhou NK, Li HD, Wang ML, Zheng J, He J, Chao WCH

EMDB-27779: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-32103: 
The unexpanded head structure of phage T4
Method: single particle / : Fang Q, Tang W, Fokine A, Mahalingam M, Shao Q, Rossmann MG, Rao VB

EMDB-32109: 
The expanded head structure of phage T4
Method: single particle / : Fang Q, Tang W, Fokine A, Mahalingam M, Shao Q, Rossmann MG, Rao VB

EMDB-32227: 
Cryo-EM structure of amyloid fibril formed by full-length human SOD1
Method: helical / : Wang LQ, Ma YY, Yuan HY, Zhao K, Zhang MY, Wang Q, Huang X, Xu WC, Chen J, Li D, Zhang DL, Zou LY, Yin P, Liu C, Liang Y

EMDB-13976: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound)
Method: single particle / : Kao WC, Hunte C

EMDB-25263: 
SARS-CoV-2 S B.1.617.2 delta variant + S2M11 + S2L20 Global Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25264: 
SARS-CoV-2 S NTD B.1.617.2 delta variant + S2L20 Local Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25265: 
SARS-CoV-2 S B.1.617.1 kappa variant + S309 + S2L20 Global Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
